I. Multiscale computational methods
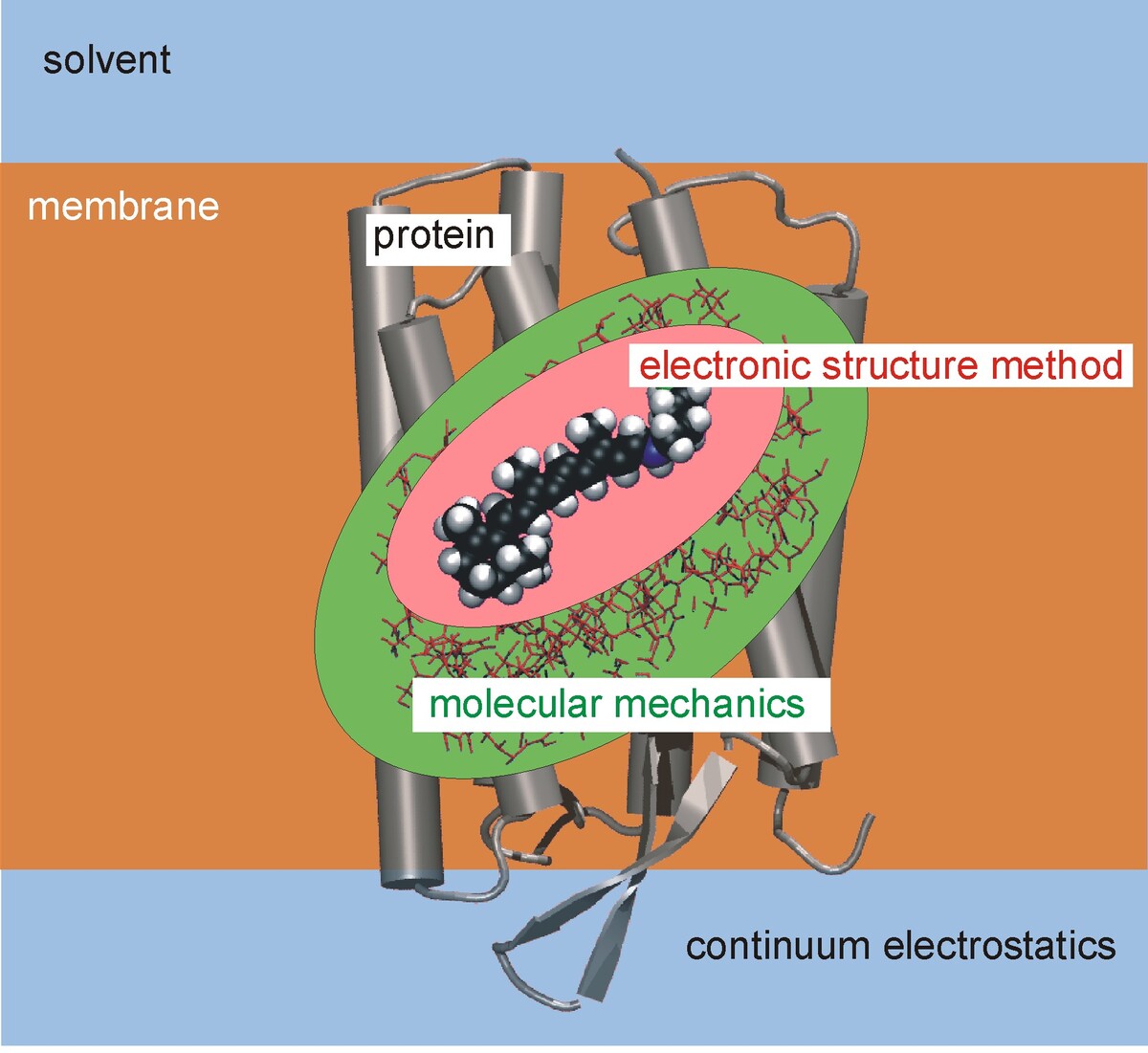
To study chemical reactivity and spectroscopic properties of complex systems like proteins or organic semi-conductors, quantum mechanical (QM) methods are required. Due to the large size of these structures, a simple application of QM methods is impossible due to the high computational cost. Therefore, QM methods are conveniently combined with empirical force field methods (Molecular Mechanics: MM) in the so- called QM/MM methods. In such approaches, the active site of a protein consisting of several tens of atoms is treated with QM, while the remainder of the protein is handled with the computationally cheaper MM methods. This approach can be taken to treat even larger scales by combining the QM/MM methods with continuum approaches. Also, various QM methods with different accuracy can be applied in the active site, building onion-like computation shells of methods.
Besides the development of such method-combinations, a large part of our effort concentrates on the development of fast and accurate approximations to Density Functional Theory (DFT) resulting in the DFTB suite of methods which are about three orders of magnitude faster than DFT, however, keeping a similar accuracy for a broad range of molecular properties.
Topics:
II. Electron Transfer in proteins and organic materials
Modeling of electron transfer (ET) events in proteins, DNA and organic materials is a particular challenge for computational chemistry due to the extensive size of the involved QM region on the one hand and to the prolonged time scales on the other. An interesting sub-class of ET processes is those which appear to be relatively fast – on the sub-nanosecond range – and for which the standard approximative schemes like Marcus' theory may no longer be valid. For such processes, we have developed a novel multi-scale methodology which intertwines efficient QM calculations with MM methods and allows for extended sampling of the processes. The electron (or electron hole) is propagated according to the Schrödinger equation within a semi-classical approach while the movement of atoms is described by classical Newton’s equations of motion.
The recent applications are concerned with several topical ET systems. In double-stranded DNA, hole transfer occurs over long distances of up to 100 ns, which has consequences for radiatively induced damage and repair processes. The mechanism of this transfer is controversial to-date, and we are performing simulations designed to resolve this issue. Another application is the hole transfer in the DNA photolyase protein, which takes place in several consecutive steps. It represents a paradigmatic system that violates the assumptions of the classical theory of ET, while our simulations describe the transfer well. A new class of systems that we are starting to work is organic semi-conductors. This project is in an early stage of development, which was made necessary by the fundamental differences between the relevant materials and the biomolecular complexes studied so far.
Topics:
III. Photo-active proteins
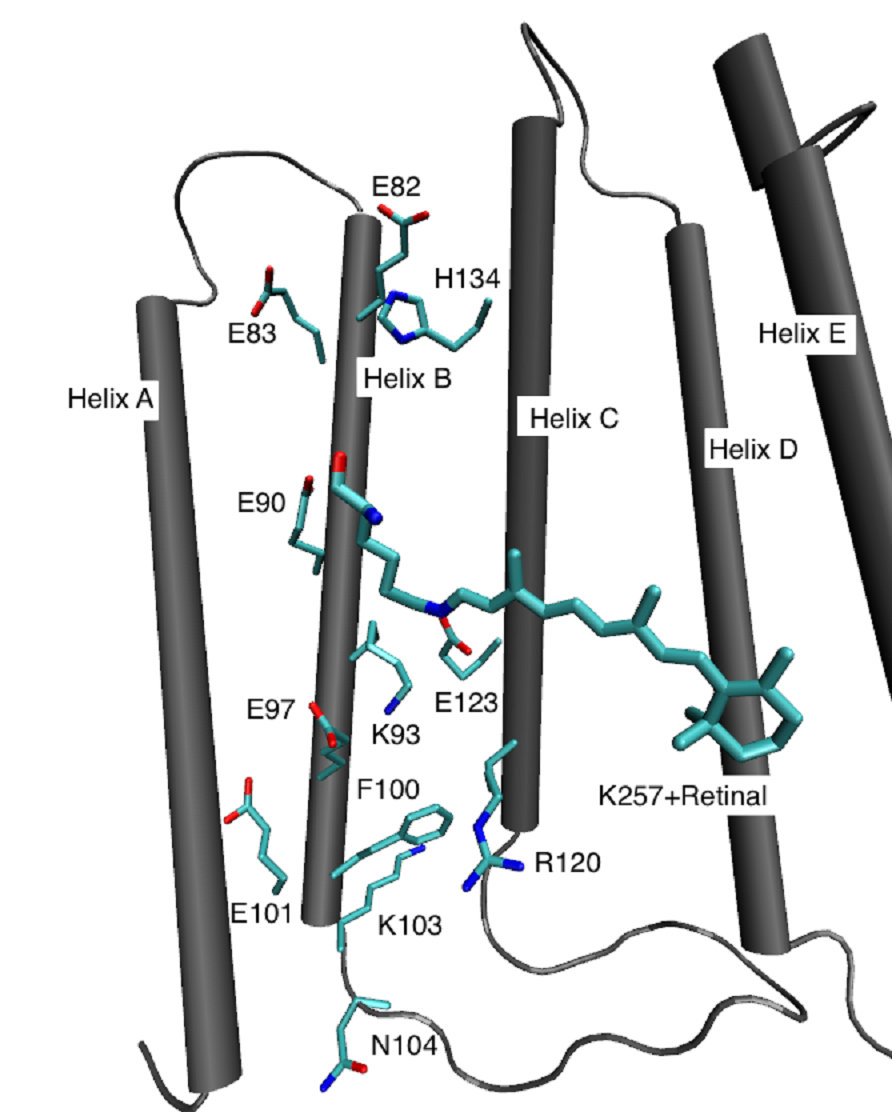
Many proteins make use of the sun light to drive important processes in the cell, like energy generation, or signal conversion. In the last years, we studied intensely the bacterial protein Bacteriorhodopsin (bR), which acts as light driven proton pump. It has been in the focus of several scientific fields for several decades, including various biological, chemical and biophysical approaches, which lead to a detailed understanding of the chemical events and structural changes that accompany the proton pumping process. Simulations, in particular those based on QM/MM methods complemented the picture by computing proton transfer pathways through the protein and energy barriers and relating spectroscopic measurements with the underlying structure.The channelrhodopsins (ChRs) are currently the most regarded members of the rhodopsin proteins and may be considered the most intriguing photo-active proteins discovered in the last decade. They are light-induced ion channels and have drawn significant attention from neuroscientists. They can be readily expressed in a variety of targets, e.g. mammalian neurons, to generate action potentials with light. The ChRs are the foundation of the new method of optogenetics. Despite of their advanced application, surprisingly little is known about their detailed structure and functional mechanism.
Topics:
Structure and Function of the Channelrhodopsines
Proton Transfer in Bacteriorhodopsin
IV. Interaction of peptides with membranes
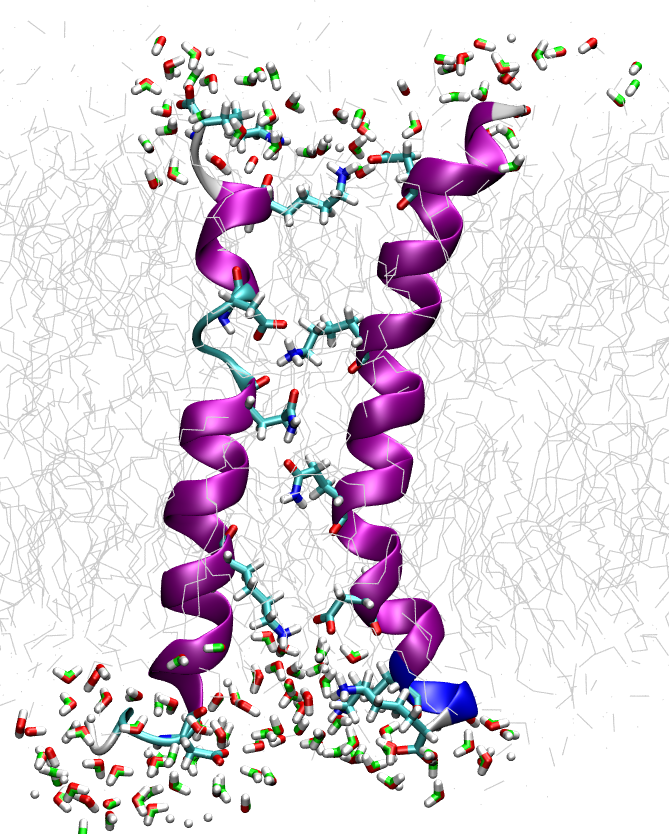
Membranes are crucial parts of every cell and a majority of biochemical reactions in metabolism, signalling and transport involves membranes and membrane proteins. Obtaining structural data for anisotropic and complex membrane systems is difficult and computer simulations can help tremendously here.
We study the physico-chemical properties of lipid bilayer biomembrane models using high-efficiency particle dynamics simulations that allow even very slow processes like phase transitions to be described. Both coarse-grained models for large scale processes and atomistic force field molecular dynamics simulations are employed. In collaboration with the group of Prof. Anne Ulrich we draw the connection between simulation results and NMR data to study the mechanism of antimicrobial, membrane penetrating peptides.